Statistics#
import sys, os
# spyder up to find the root
oceanbench_root = "/gpfswork/rech/cli/uvo53rl/projects/oceanbench"
# append to path
sys.path.append(str(oceanbench_root))
import autoroot
import typing as tp
import jax
import jax.numpy as jnp
import jax.scipy as jsp
import jax.random as jrandom
import numpy as np
import numba as nb
import pandas as pd
import equinox as eqx
import kernex as kex
import finitediffx as fdx
import diffrax as dfx
import xarray as xr
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm.notebook import tqdm, trange
from jaxtyping import Float, Array, PyTree, ArrayLike
import wandb
from omegaconf import OmegaConf
import hydra
import metpy
from sklearn.pipeline import Pipeline
from jejeqx._src.transforms.dataframe.spatial import Spherical2Cartesian
from jejeqx._src.transforms.dataframe.temporal import TimeDelta
from jejeqx._src.transforms.dataframe.scaling import MinMaxDF
sns.reset_defaults()
sns.set_context(context="poster", font_scale=0.7)
jax.config.update("jax_enable_x64", False)
%matplotlib inline
%load_ext autoreload
%autoreload 2
Processing Chain#
Part I:
Open Dataset
Validate Coordinates + Variables
Decode Time
Select Region
Sortby Time
Part II: Regrid
Part III:
Interpolate Nans
Add Units
Spatial Rescale
Time Rescale
Part IV: Metrics
Data#
# !wget wget -nc https://s3.us-east-1.wasabisys.com/melody/osse_data/ref/NATL60-CJM165_GULFSTREAM_ssh_y2013.1y.nc
!ls /gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/staging/results/DUACS
# !cat configs/postprocess.yaml
# # load config
# config_dm = OmegaConf.load('./configs/postprocess.yaml')
# # instantiate
# ds = hydra.utils.instantiate(config_dm.NATL60_GF_1Y1D)
# ds
from oceanbench._src.geoprocessing.gridding import (
grid_to_regular_grid,
coord_based_to_grid,
)
from oceanbench._src.geoprocessing import geostrophic as geocalc
from metpy.units import units
def calculate_physical_quantities(da):
da["ssh"] = da.ssh * units.meters
da = geocalc.streamfunction(da, "ssh")
da = geocalc.geostrophic_velocities(da, variable="psi")
da = geocalc.kinetic_energy(da, variables=["u", "v"])
da = geocalc.divergence(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, "div")
da = geocalc.relative_vorticity(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, "vort_r")
da = geocalc.strain_magnitude(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, variable="strain")
return da
def correct_labels(ds):
ds["lon"].attrs["units"] = "degrees"
ds["lat"].attrs["units"] = "degrees"
ds["ssh"].attrs["units"] = "m"
ds["ssh"].attrs["standard_name"] = "sea_surface_height"
ds["ssh"].attrs["long_name"] = "Sea Surface Height"
ds["lon"].attrs["standard_name"] = "longitude"
ds["lat"].attrs["standard_name"] = "latitude"
ds["lat"].attrs["long_name"] = "Latitude"
ds["lon"].attrs["long_name"] = "Longitude"
return ds
Reference Dataset#
For the reference dataset, we will look at the NEMO simulation of the Gulfstream.
%%time
# load config
config_dm = OmegaConf.load("./configs/postprocess.yaml")
# instantiate
ds_natl60 = hydra.utils.instantiate(config_dm.NATL60_GF_FULL).compute()
ds_natl60
CPU times: user 12.3 s, sys: 2.6 s, total: 14.9 s
Wall time: 47.8 s
<xarray.Dataset>
Dimensions: (time: 42, lat: 600, lon: 600)
Coordinates:
* lon (lon) float64 -64.98 -64.97 -64.95 -64.93 ... -55.03 -55.02 -55.0
* lat (lat) float64 33.02 33.03 33.05 33.07 ... 42.95 42.97 42.98 43.0
* time (time) datetime64[ns] 2012-10-22 2012-10-23 ... 2012-12-02
Data variables:
ssh (time, lat, lon) float32 0.6549 0.6571 0.6593 ... -0.2152 -0.2174
Attributes:
Info: Horizontal grid read in regulargrid_NATL60.nc / Source field re...
About: Created by SOSIE interpolation environement => https://github.c...AlongTrack -> Uniform Grid#
# load config
psd_config = OmegaConf.load("./configs/metrics.yaml")
ds_natl60 = hydra.utils.instantiate(psd_config.fill_nans)(ds_natl60.pint.dequantify())
Coarsend Versions#
ds_natl60 = ds_natl60.coarsen({"lon": 3, "lat": 3}).mean()
ds_natl60
<xarray.Dataset>
Dimensions: (time: 42, lat: 200, lon: 200)
Coordinates:
* lon (lon) float64 -64.97 -64.92 -64.87 -64.82 ... -55.12 -55.07 -55.02
* lat (lat) float64 33.03 33.08 33.13 33.18 ... 42.83 42.88 42.93 42.98
* time (time) datetime64[ns] 2012-10-22 2012-10-23 ... 2012-12-02
Data variables:
ssh (time, lat, lon) float32 0.652 0.6585 0.6642 ... -0.2079 -0.2149
Attributes:
Info: Horizontal grid read in regulargrid_NATL60.nc / Source field re...
About: Created by SOSIE interpolation environement => https://github.c...Prediction Datasets#
%%time
# load config
experiment = "swot" # "nadir" #
if experiment == "nadir":
# load config
results_config = OmegaConf.load(f"./configs/results_dc20a_nadir.yaml")
# instantiate
ds_duacs = hydra.utils.instantiate(results_config.DUACS_NADIR.data).compute()
ds_miost = hydra.utils.instantiate(results_config.MIOST_NADIR.data).compute()
ds_nerf_siren = hydra.utils.instantiate(
results_config.NERF_SIREN_NADIR.data
).compute()
ds_nerf_ffn = hydra.utils.instantiate(results_config.NERF_FFN_NADIR.data).compute()
ds_nerf_mlp = hydra.utils.instantiate(results_config.NERF_MLP_NADIR.data).compute()
elif experiment == "swot":
# load config
results_config = OmegaConf.load(f"./configs/results_dc20a_swot.yaml")
# instantiate
ds_duacs = hydra.utils.instantiate(results_config.DUACS_SWOT.data).compute()
ds_miost = hydra.utils.instantiate(results_config.MIOST_SWOT.data).compute()
ds_nerf_siren = hydra.utils.instantiate(
results_config.NERF_SIREN_SWOT.data
).compute()
ds_nerf_ffn = hydra.utils.instantiate(results_config.NERF_FFN_SWOT.data).compute()
ds_nerf_mlp = hydra.utils.instantiate(results_config.NERF_MLP_SWOT.data).compute()
CPU times: user 174 ms, sys: 41.8 ms, total: 216 ms
Wall time: 497 ms
!ls /gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/staging/ml_ready/
Regrdding#
Uniform Grid –> Uniform Grid#
%%time
ds_duacs = grid_to_regular_grid(
src_grid_ds=ds_duacs.pint.dequantify(),
tgt_grid_ds=ds_natl60.pint.dequantify(),
keep_attrs=False,
)
ds_miost = grid_to_regular_grid(
src_grid_ds=ds_miost.pint.dequantify(),
tgt_grid_ds=ds_natl60.pint.dequantify(),
keep_attrs=False,
)
ds_nerf_siren = grid_to_regular_grid(
src_grid_ds=ds_nerf_siren.pint.dequantify(),
tgt_grid_ds=ds_natl60.pint.dequantify(),
keep_attrs=False,
)
ds_nerf_ffn = grid_to_regular_grid(
src_grid_ds=ds_nerf_ffn.pint.dequantify(),
tgt_grid_ds=ds_natl60.pint.dequantify(),
keep_attrs=False,
)
ds_nerf_mlp = grid_to_regular_grid(
src_grid_ds=ds_nerf_mlp.pint.dequantify(),
tgt_grid_ds=ds_natl60.pint.dequantify(),
keep_attrs=False,
)
CPU times: user 10.2 s, sys: 93.1 ms, total: 10.3 s
Wall time: 10.3 s
Preprocess Chain#
%%time
# load config
psd_config = OmegaConf.load("./configs/metrics.yaml")
ds_duacs = hydra.utils.instantiate(psd_config.fill_nans)(ds_duacs.pint.dequantify())
ds_miost = hydra.utils.instantiate(psd_config.fill_nans)(ds_miost.pint.dequantify())
ds_nerf_siren = hydra.utils.instantiate(psd_config.fill_nans)(
ds_nerf_siren.pint.dequantify()
)
ds_nerf_ffn = hydra.utils.instantiate(psd_config.fill_nans)(
ds_nerf_ffn.pint.dequantify()
)
ds_nerf_mlp = hydra.utils.instantiate(psd_config.fill_nans)(
ds_nerf_mlp.pint.dequantify()
)
CPU times: user 987 ms, sys: 2.01 ms, total: 989 ms
Wall time: 992 ms
Geophysical Variables#
def calculate_physical_quantities(da):
da["ssh"] = da.ssh * units.meters
da = geocalc.streamfunction(da, "ssh")
da = geocalc.geostrophic_velocities(da, variable="psi")
da = geocalc.kinetic_energy(da, variables=["u", "v"])
da = geocalc.divergence(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, "div")
da = geocalc.relative_vorticity(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, "vort_r")
da = geocalc.strain_magnitude(da, variables=["u", "v"])
da = geocalc.coriolis_normalized(da, variable="strain")
return da
%%time
ds_natl60 = calculate_physical_quantities(correct_labels(ds_natl60).pint.dequantify())
ds_duacs = calculate_physical_quantities(correct_labels(ds_duacs).pint.dequantify())
ds_miost = calculate_physical_quantities(correct_labels(ds_miost).pint.dequantify())
ds_nerf_siren = calculate_physical_quantities(
correct_labels(ds_nerf_siren).pint.dequantify()
)
ds_nerf_ffn = calculate_physical_quantities(
correct_labels(ds_nerf_ffn).pint.dequantify()
)
ds_nerf_mlp = calculate_physical_quantities(
correct_labels(ds_nerf_mlp).pint.dequantify()
)
CPU times: user 3.39 s, sys: 332 ms, total: 3.72 s
Wall time: 3.74 s
Absolute Statistics#
from matplotlib import ticker
import xskillscore
from oceanbench._src.metrics.stats import nrmse_da, rmse_da
def absolute_error_plot(da, da_ref, variable="ssh", color="red"):
r2 = xskillscore.r2(
da[variable].pint.dequantify(),
da_ref[variable].pint.dequantify(),
dim=["lat", "lon", "time"],
)
slope = xskillscore.linslope(
da[variable].pint.dequantify(),
da_ref[variable].pint.dequantify(),
dim=["lat", "lon", "time"],
)
nrmse = nrmse_da(
da=da, da_ref=da_ref, variable=variable, dim=["lon", "lat", "time"]
)
nrmse_std = nrmse_da(
da=da, da_ref=da_ref, variable=variable, dim=["lon", "lat"]
).std()
from matplotlib.offsetbox import AnchoredText
fig, ax = plt.subplots(figsize=(7, 7))
id_line = np.linspace(-0.75, 1.25)
xmin = 1.2 * da_ref[variable].min()
xmax = 1.05 * da_ref[variable].max()
ax.scatter(
x=da[variable].values.ravel(),
y=da_ref[variable].values.ravel(),
s=0.1,
alpha=0.1,
color=color,
)
ax.plot(id_line, id_line, color="black", zorder=2, linewidth=5)
ax.set(
xlim=[xmin, xmax],
ylim=[xmin, xmax],
xlabel="Predictions",
ylabel="Ground Truth",
)
# ax.autoscale(enable=True, axis='both', tight=True)
ax.set_aspect("equal")
at = AnchoredText(
f"R$^2$: {r2.values:.3f}" +
f"\nSlope: {slope.values:.2f}"+
f"\nnRMSE($\mu$): {nrmse.values:.2f}"+
f"\nnRMSE($\sigma$): {nrmse_std.values:.2f}",
prop=dict(fontsize=16),
frameon=True,
loc="upper left",
)
at.patch.set_boxstyle("round,pad=0.,rounding_size=0.2")
ax.add_artist(at)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.xaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.1f}"))
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.1f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.25))
ax.xaxis.set_major_locator(ticker.MultipleLocator(0.25))
plt.tight_layout()
return fig, ax
# DUACS
fig, ax = absolute_error_plot(ds_duacs, ds_natl60, "ssh", color="tab:green")
fig.savefig(f"./figures/dc20a/stats/error_duacs_{experiment}.png")
# MIOST
fig, ax = absolute_error_plot(ds_miost, ds_natl60, "ssh", color="tab:red")
fig.savefig(f"./figures/dc20a/stats/error_miost_{experiment}.png")
# NERF (MLP)
fig, ax = absolute_error_plot(ds_nerf_mlp, ds_natl60, "ssh", color="tab:olive")
fig.savefig(f"./figures/dc20a/stats/error_nerf_mlp_{experiment}.png")
# NERF (FFN)
fig, ax = absolute_error_plot(ds_nerf_ffn, ds_natl60, "ssh", color="tab:blue")
fig.savefig(f"./figures/dc20a/stats/error_nerf_ffn_{experiment}.png")
# NERF (SIREN)
fig, ax = absolute_error_plot(ds_nerf_siren, ds_natl60, "ssh", color="tab:cyan")
fig.savefig(f"./figures/dc20a/stats/error_nerf_siren_{experiment}.png")
Temporal Statistics#
ds_duacs_nrmse = rmse_da(
da=ds_duacs, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_miost_nrmse = rmse_da(
da=ds_miost, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_mlp_nrmse = rmse_da(
da=ds_nerf_mlp, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_ffn_nrmse = rmse_da(
da=ds_nerf_ffn, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_siren_nrmse = rmse_da(
da=ds_nerf_siren, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_nrmse.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_nrmse.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_nrmse.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_nrmse.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_nrmse.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.01, 0.100], ylabel="RMSE [m]", xlabel="Date")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.02))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/rmse_space_{experiment}.png")
plt.show()
ds_duacs_nrmse = nrmse_da(
da=ds_duacs, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_miost_nrmse = nrmse_da(
da=ds_miost, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_mlp_nrmse = nrmse_da(
da=ds_nerf_mlp, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_ffn_nrmse = nrmse_da(
da=ds_nerf_ffn, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
ds_nerf_siren_nrmse = nrmse_da(
da=ds_nerf_siren, da_ref=ds_natl60, variable="ssh", dim=["lon", "lat"]
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_nrmse.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_nrmse.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_nrmse.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_nrmse.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_nrmse.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.5, 1.0], ylabel="Normalized RMSE", xlabel="Date")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.1f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.1))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.05))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/nrmse_space_{experiment}.png")
plt.show()
ds_duacs_nrmse = nrmse_da(
da=ds_duacs, da_ref=ds_natl60, variable="ssh", dim=["time", "lon"]
)
ds_miost_nrmse = nrmse_da(
da=ds_miost, da_ref=ds_natl60, variable="ssh", dim=["time", "lon"]
)
ds_nerf_mlp_nrmse = nrmse_da(
da=ds_nerf_mlp, da_ref=ds_natl60, variable="ssh", dim=["time", "lon"]
)
ds_nerf_ffn_nrmse = nrmse_da(
da=ds_nerf_ffn, da_ref=ds_natl60, variable="ssh", dim=["time", "lon"]
)
ds_nerf_siren_nrmse = nrmse_da(
da=ds_nerf_siren, da_ref=ds_natl60, variable="ssh", dim=["time", "lon"]
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_nrmse.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_nrmse.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_nrmse.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_nrmse.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_nrmse.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.5, 1.0], ylabel="Normalized RMSE", xlabel="Latitude [degrees]")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.1f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.1))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.05))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/nrmse_spacetime_{experiment}.png")
plt.show()
Higher Level Statistics#
from hyppo.independence import RV, Dcorr, Hsic
from hyppo.d_variate import dHsic
from hyppo.ksample import Energy
import xskillscore
def hyppo_fn_pixel(f):
def fn(x, y):
return f(x.flatten()[:, None], y.flatten()[:, None])
return fn
def hyppo_fn_2D(f):
def fn(x, y):
return f(x, y)
return fn
Pearson#
Temporal#
ds_duacs_pearsonr = xskillscore.pearson_r(
ds_duacs.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["lat", "lon"]
)
ds_miost_pearsonr = xskillscore.pearson_r(
ds_miost.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["lat", "lon"]
)
ds_nerf_mlp_pearsonr = xskillscore.pearson_r(
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
ds_nerf_ffn_pearsonr = xskillscore.pearson_r(
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
ds_nerf_siren_pearsonr = xskillscore.pearson_r(
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_pearsonr.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_pearsonr.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_pearsonr.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_pearsonr.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_pearsonr.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
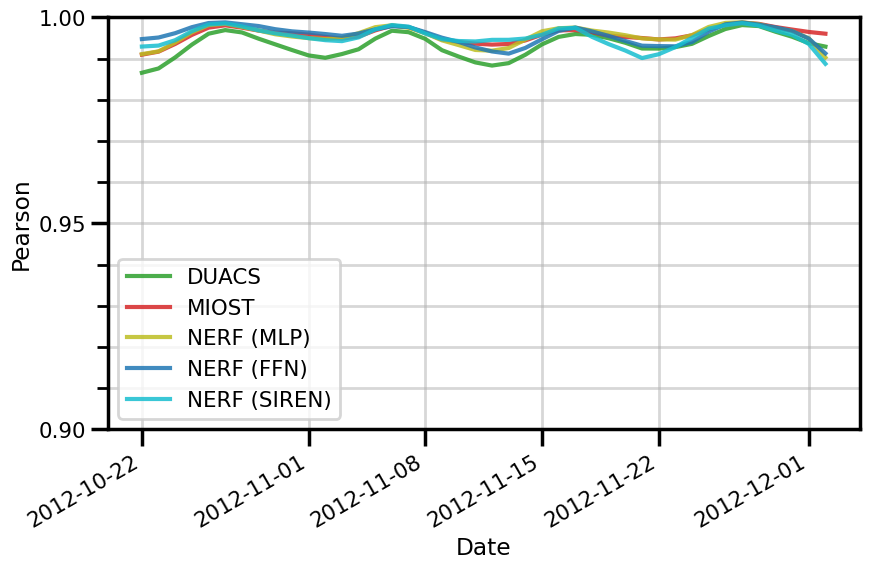
ax.set(
ylim=[0.90, 1.0],
ylabel="Pearson",
xlabel="Date"
)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/pearson_space_{experiment}.png")
plt.show()
ds_duacs_pearsonr = xskillscore.pearson_r(
ds_duacs.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["time", "lon"]
)
ds_miost_pearsonr = xskillscore.pearson_r(
ds_miost.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["time", "lon"]
)
ds_nerf_mlp_pearsonr = xskillscore.pearson_r(
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
ds_nerf_ffn_pearsonr = xskillscore.pearson_r(
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
ds_nerf_siren_pearsonr = xskillscore.pearson_r(
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_pearsonr.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_pearsonr.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_pearsonr.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_pearsonr.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_pearsonr.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.75, 1.0], ylabel="Pearson", xlabel="Latitude [degrees]")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/nrmse_spacetime_{experiment}.png")
plt.show()
Spearman#
Temporal#
ds_duacs_pearsonr = xskillscore.spearman_r(
ds_duacs.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["lat", "lon"]
)
ds_miost_pearsonr = xskillscore.spearman_r(
ds_miost.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["lat", "lon"]
)
ds_nerf_mlp_pearsonr = xskillscore.spearman_r(
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
ds_nerf_ffn_pearsonr = xskillscore.spearman_r(
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
ds_nerf_siren_pearsonr = xskillscore.spearman_r(
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["lat", "lon"],
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_pearsonr.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_pearsonr.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_pearsonr.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_pearsonr.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_pearsonr.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
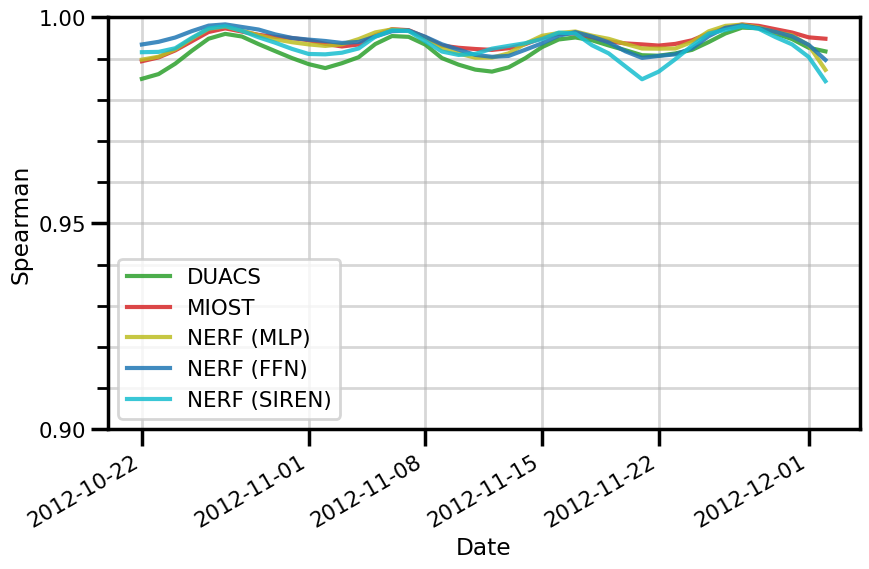
ax.set(
ylim=[0.90, 1.0],
ylabel="Spearman",
xlabel="Date"
)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/spearman_space_{experiment}.png")
plt.show()
ds_duacs_pearsonr = xskillscore.spearman_r(
ds_duacs.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["time", "lon"]
)
ds_miost_pearsonr = xskillscore.spearman_r(
ds_miost.ssh.pint.dequantify(), ds_natl60.ssh.pint.dequantify(), dim=["time", "lon"]
)
ds_nerf_mlp_pearsonr = xskillscore.spearman_r(
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
ds_nerf_ffn_pearsonr = xskillscore.spearman_r(
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
ds_nerf_siren_pearsonr = xskillscore.spearman_r(
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
dim=["time", "lon"],
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_pearsonr.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_pearsonr.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_pearsonr.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_pearsonr.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_pearsonr.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
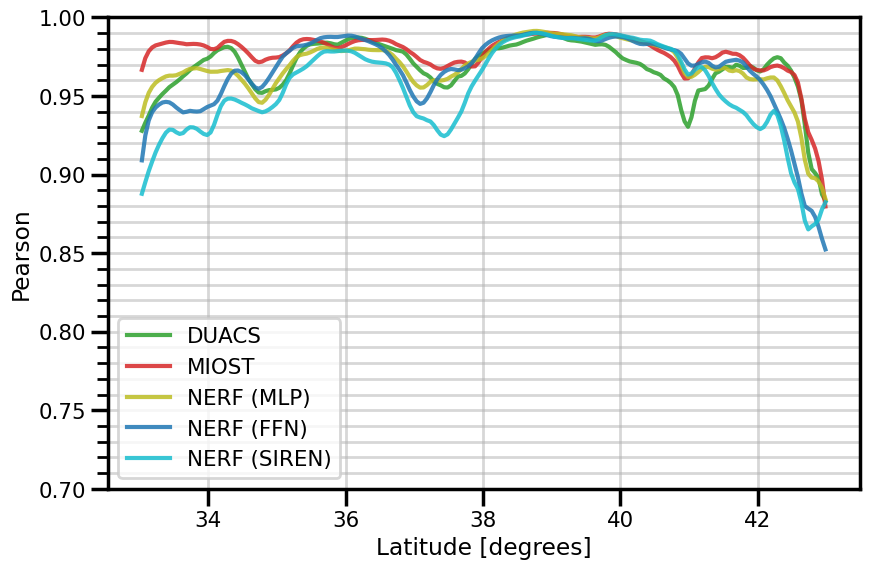
ax.set(
ylim=[0.70, 1.0],
ylabel="Pearson",
xlabel="Latitude [degrees]"
)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/spearman_spacetime_{experiment}.png")
plt.show()
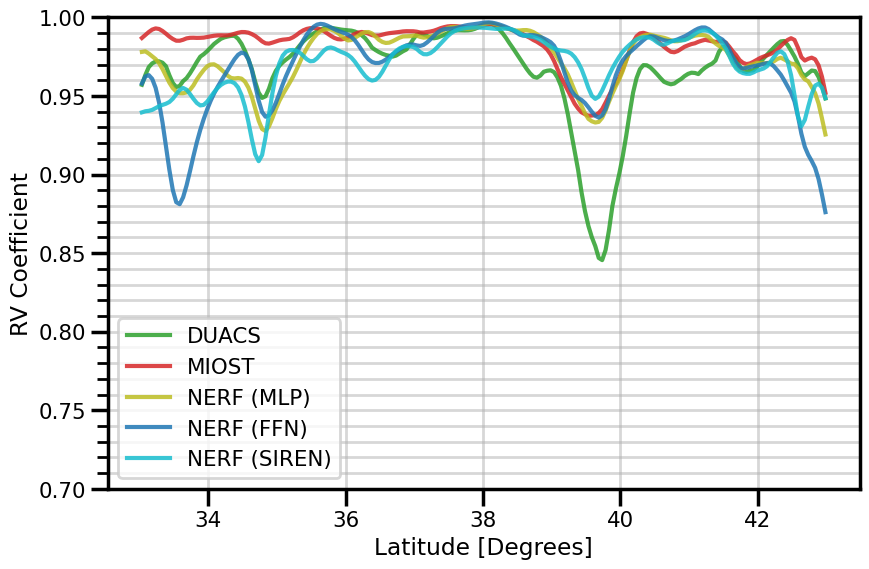
RV Coefficient#
def rv_coefficient(x, y):
return RV().statistic(x, y)
ds_duacs_rv = xr.apply_ufunc(
rv_coefficient,
ds_duacs.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_miost_rv = xr.apply_ufunc(
rv_coefficient,
ds_miost.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_mlp_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_ffn_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_siren_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_rv.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_rv.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_rv.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_rv.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_rv.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.90, 1.0], ylabel="RV Coefficient", xlabel="Date")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/rv_space_{experiment}.png")
plt.show()
ds_duacs_rv = xr.apply_ufunc(
rv_coefficient,
ds_duacs.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_miost_rv = xr.apply_ufunc(
rv_coefficient,
ds_miost.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_mlp_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_ffn_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_siren_rv = xr.apply_ufunc(
rv_coefficient,
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_rv.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_rv.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_rv.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_rv.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_rv.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(
ylim=[0.70, 1.0],
ylabel="RV Coefficient",
xlabel="Latitude [Degrees]"
)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/rv_spacetime_{experiment}.png")
plt.show()
CKA - Linear#
def cka_linear(x, y):
xy = dHsic(compute_kernel="linear").statistic(x, y)
xx = dHsic(compute_kernel="linear").statistic(x, x)
yy = dHsic(compute_kernel="linear").statistic(y, y)
return xy / (np.sqrt(xx) * np.sqrt(yy))
ds_duacs_rv = xr.apply_ufunc(
cka_linear,
ds_duacs.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_miost_rv = xr.apply_ufunc(
cka_linear,
ds_miost.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_mlp_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_ffn_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_siren_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["lat", "lon"], ["lat", "lon"]],
exclude_dims={"lat", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_rv.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_rv.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_rv.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_rv.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_rv.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
ax.set(ylim=[0.90, 1.0], ylabel="CKA (Linear)", xlabel="Date")
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/cka_linear_space_{experiment}.png")
plt.show()
ds_duacs_rv = xr.apply_ufunc(
cka_linear,
ds_duacs.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_miost_rv = xr.apply_ufunc(
cka_linear,
ds_miost.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_mlp_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_mlp.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_ffn_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_ffn.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
ds_nerf_siren_rv = xr.apply_ufunc(
cka_linear,
ds_nerf_siren.ssh.pint.dequantify(),
ds_natl60.ssh.pint.dequantify(),
input_core_dims=[["time", "lon"], ["time", "lon"]],
exclude_dims={"time", "lon"},
output_dtypes=[np.float64],
vectorize=True,
)
fig, ax = plt.subplots(figsize=(9, 6))
ds_duacs_rv.plot(ax=ax, label="DUACS", color="tab:green", alpha=0.85)
ds_miost_rv.plot(ax=ax, label="MIOST", color="tab:red", alpha=0.85)
ds_nerf_mlp_rv.plot(ax=ax, label="NERF (MLP)", color="tab:olive", alpha=0.85)
ds_nerf_ffn_rv.plot(ax=ax, label="NERF (FFN)", color="tab:blue", alpha=0.85)
ds_nerf_siren_rv.plot(ax=ax, label="NERF (SIREN)", color="tab:cyan", alpha=0.85)
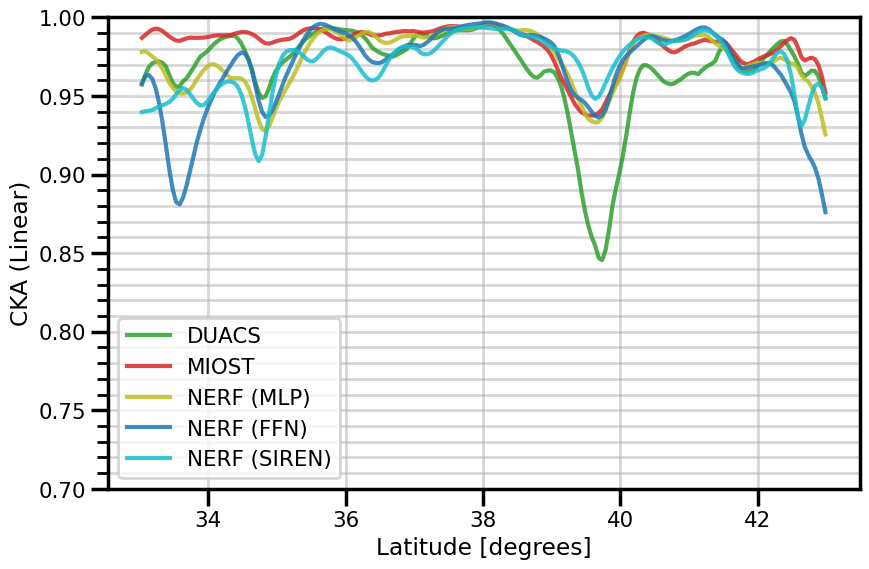
ax.set(
ylim=[0.70, 1.0],
ylabel="CKA (Linear)",
xlabel="Latitude [degrees]"
)
ax.grid("on", which="both", axis="both", alpha=0.5)
# tick format
ax.yaxis.set_major_formatter(ticker.StrMethodFormatter("{x:.2f}"))
# tick locator
ax.yaxis.set_major_locator(ticker.MultipleLocator(0.05))
ax.yaxis.set_minor_locator(ticker.MultipleLocator(0.01))
plt.tight_layout()
plt.legend()
fig.savefig(f"./figures/dc20a/stats/cka_spacetime_{experiment}.png")
plt.show()