Gradient Considerations#
import autoroot
import xarray as xr
xrds = xr.open_dataset("/Volumes/EMANS_HDD/data/oceanbench-data-registry/osse_natl60/grid/gf_obs_nadir.nc")
xrds
<xarray.Dataset>
Dimensions: (lat: 201, lon: 201, time: 365)
Coordinates:
* lon (lon) float64 -65.0 -64.95 -64.9 -64.85 ... -55.1 -55.05 -55.0
* lat (lat) float64 33.0 33.05 33.1 33.15 ... 42.85 42.9 42.95 43.0
* time (time) datetime64[ns] 2012-10-01 2012-10-02 ... 2013-09-30
Data variables:
mask (lat, lon) float64 ...
lag (time, lat, lon) float64 ...
flag (time, lat, lon) float64 ...
ssh_obs (time, lat, lon) float64 ...
ssh_mod (time, lat, lon) float64 ...
anomaly_obs (time, lat, lon) float64 ...
anomaly_mod (time, lat, lon) float64 ...import autoroot
import typing as tp
import jax
import jax.numpy as jnp
import jax.scipy as jsp
import jax.random as jrandom
import numpy as np
import numba as nb
import pandas as pd
import equinox as eqx
import kernex as kex
import finitediffx as fdx
import diffrax as dfx
import xarray as xr
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm.notebook import tqdm, trange
from jaxtyping import Float, Array, PyTree, ArrayLike
import wandb
from omegaconf import OmegaConf
import hydra
import metpy
from sklearn.pipeline import Pipeline
from jejeqx._src.transforms.dataframe.spatial import Spherical2Cartesian
from jejeqx._src.transforms.dataframe.temporal import TimeDelta
from jejeqx._src.transforms.dataframe.scaling import MinMaxDF
sns.reset_defaults()
sns.set_context(context="talk", font_scale=0.7)
jax.config.update("jax_enable_x64", False)
%matplotlib inline
%load_ext autoreload
%autoreload 2
Recap Formulation#
We are interested in learning non-linear functions \(\boldsymbol{f}\).
where the \(\boldsymbol{\phi}(\cdot)\) is a basis function. Neural Fields typically try to learn this basis funciton via a series of composite functions of the form
Problems#
Here, we will demonstrate a very simple spatial interpolation problem for sea surface height.
Data#
# !wget wget -nc https://s3.us-east-1.wasabisys.com/melody/osse_data/ref/NATL60-CJM165_GULFSTREAM_ssh_y2013.1y.nc
from pathlib import Path
Path(
"/gpfswork/rech/cli/uvo53rl/projects/jejeqx/data/natl60/NATL60-CJM165_GULFSTREAM_ssh_y2013.1y.nc"
).is_file()
!ls configs
!pwd
eval_natl60_dc20a.yaml natl60_obs.yaml results_dc20a_nadir.yaml
lr_scheduler.yaml natl60.yaml results_dc20a_swot.yaml
metrics.yaml optimizer.yaml
model.yaml postprocess.yaml
/gpfsdswork/projects/rech/cli/uvo53rl/projects/jejeqx/notebooks/dev/nerfs
!cat configs/natl60.yaml
natl60_dc20a:
_target_: jejeqx._src.datamodules.coords_v2.AlongTrackDM
paths: "/gpfswork/rech/cli/uvo53rl/projects/jejeqx/data/natl60/NATL60-CJM165_GULFSTREAM_ssh_y2013.1y.nc"
batch_size: 10_000
shuffle: True
train_size: 0.80
subset_size: 0.40
spatial_coords: ["lat", "lon"]
temporal_coords: ["time"]
variables: ["ssh"]
evaluation: False
resample: "1D"
decode_times: False
spatial_units: "meters"
time_unit: "seconds"
time_freq: 1
t0: "2013-01-01"
select:
time: {_target_: builtins.slice, _args_: ["2013-01-01", "2013-01-01"]}
natl60_dc20a_scaled:
_target_: jejeqx._src.datamodules.coords_v2.AlongTrackDM
paths: "/gpfswork/rech/cli/uvo53rl/projects/jejeqx/data/natl60/NATL60-CJM165_GULFSTREAM_ssh_y2013.1y.nc"
batch_size: 10_000
shuffle: True
train_size: 0.80
subset_size: 0.40
spatial_coords: ["lat", "lon"]
temporal_coords: ["time"]
variables: ["ssh"]
evaluation: False
resample: "1D"
decode_times: False
spatial_units: "meters"
time_unit: "seconds"
time_freq: 1
t0: "2013-01-01"
select:
time: {_target_: builtins.slice, _args_: ["2013-01-01", "2013-01-01"]}
temporal_transform: ${temporal_transforms}
spatial_transform: ${spatial_transforms}
spatial_transforms:
_target_: jejeqx._src.transforms.pipelines.make_pipeline
_recursive_: False
steps_config:
- spatialminmax:
_target_: jejeqx._src.transforms.dataframe.scaling.MinMaxDF
columns: ["lat", "lon"]
min_val: -1
max_val: 1
temporal_transforms:
_target_: jejeqx._src.transforms.pipelines.make_pipeline
_recursive_: False
steps_config:
- timeminmax:
_target_: jejeqx._src.transforms.dataframe.scaling.MinMaxDF
columns: ["time"]
min_val: -1
max_val: 1
natl60_dc20a_eval:
_target_: "jejeqx._src.datamodules.coords.EvalCoordDM"
paths: "/gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/test/dc_ref/NATL60-CJM165_GULFSTREAM*"
batch_size: 10_000
shuffle: False
train_size: 0.80
decode_times: True
evaluation: True
spatial_coords: ["lat", "lon"]
temporal_coords: ["time"]
variables: ["sossheig"]
coarsen:
lon: 2
lat: 2
resample: "1D"
select:
time: {_target_: builtins.slice, _args_: ["2012-10-01", "2012-12-02"]}
# !cat configs/natl60.yaml
# load config
config_dm = OmegaConf.load('/gpfsdswork/projects/rech/cli/uvo53rl/projects/jejeqx/notebooks/dev/nerfs/configs/natl60.yaml')
# instantiate
dm = hydra.utils.instantiate(config_dm.natl60_dc20a)
# run setup
dm.setup()
# check cunits
(
dm.ds_test[:]["spatial"].min(),
dm.ds_test[:]["spatial"].max(),
dm.ds_test[:]["temporal"].min(),
dm.ds_test[:]["temporal"].max(),
dm.ds_test[:]["data"].min(),
dm.ds_test[:]["data"].max(),
)
(0.0, 1111948.7428468189, 0.0, 0.0, -0.46802905337971806, 1.0029388256616805)
# spatial transform
spatial_transforms = hydra.utils.instantiate(config_dm.spatial_transforms)
spatial_transforms
temporal_transforms = hydra.utils.instantiate(config_dm.temporal_transforms)
temporal_transforms
Pipeline(steps=[('timeminmax', MinMaxDF(columns=['time']))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('timeminmax', MinMaxDF(columns=['time']))])MinMaxDF(columns=['time'])
# instantiate
dm = hydra.utils.instantiate(
config_dm.natl60_dc20a_scaled,
)
# run setup
dm.setup()
# check units
(
dm.ds_test[:]["spatial"].min(),
dm.ds_test[:]["spatial"].max(),
dm.ds_test[:]["temporal"].min(),
dm.ds_test[:]["temporal"].max(),
dm.ds_test[:]["data"].min(),
dm.ds_test[:]["data"].max(),
)
(-1.0, 1.0, -1.0, -1.0, -0.46802905337971806, 1.0029388256616805)
len(dm.ds_train)
12928
xrda = dm.load_xrds()
print(f"Field Shape: {xrda.ssh.shape}")
print(f"Number of Coords: {len(dm.ds_train):,}")
from matplotlib import ticker
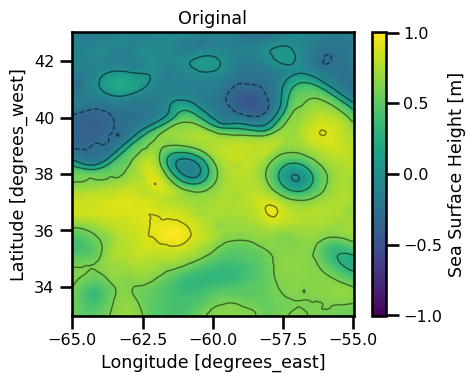
fig, ax = plt.subplots(ncols=1, figsize=(5,4))
subset_ds = xrda.ssh.isel(time=0)
cbar_kwargs = {"label": "Sea Surface Height [m]"}
subset_ds.plot.pcolormesh(ax=ax, cmap="viridis", cbar_kwargs=cbar_kwargs)
loc = ticker.MaxNLocator(5)
levels = loc.tick_values(xrda.ssh.min().values, xrda.ssh.max().values)
subset_ds.plot.contour(
ax=ax,
alpha=0.5, linewidths=1, cmap="black",
levels=levels,
linestyles=np.where(levels >= 0, "-", "--")
# vmin=vmin, vmax=vmax,
# **kwargs
)
ax.set(title="Original")
plt.tight_layout()
plt.show()
init = dm.ds_train[:32]
x_init, t_init, y_init = init["spatial"], init["temporal"], init["data"]
x_init.min(), x_init.max(), x_init.shape, t_init.min(), t_init.max(), t_init.shape
(-0.98, 1.0, (32, 2), -1.0, -1.0, (32, 1))
Model#
The input data is a coordinate vector, \(\mathbf{x}_\phi\), of the image coordinates.
where \(D_\phi = [\text{x}, \text{y}]\). So we are interested in learning a function, \(\boldsymbol{f}\), such that we can input a coordinate vector and output a scaler/vector value of the pixel value.
MLP Layer#
where \(\sigma\) is the ReLU activation function.
!cat ./configs/model.yaml
# load config
model_config = OmegaConf.load("./configs/model.yaml")
# instantiate
model_mlp = hydra.utils.instantiate(model_config.mlp)
# test output
out = model_mlp(x=x_init[0], t=t_init[0])
assert out.shape == y_init[0].shape
# test output (batched)
out_batch = jax.vmap(model_mlp, in_axes=(0, 0))(x_init, t_init)
assert out_batch.shape == y_init.shape
No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
Optimizer (+ Learning Rate)#
For this, we will use a simple adam optimizer with a learning_rate of 1e-4. From many studies, it appears that a lower learning rate works well with this methods because there is a lot of data. In addition, a bigger batch_size is also desireable. We will set the num_epochs to 2_000 which should be good enough for a single image. Obviously more epochs and a better learning rate scheduler would result in better results but this will be sufficient for this demo.
import optax
num_epochs = 5000
# load config
opt_config = OmegaConf.load("./configs/optimizer.yaml")
# instantiate
optimizer = hydra.utils.instantiate(opt_config.adamw)
scheduler_config = OmegaConf.load("./configs/lr_scheduler.yaml")
num_steps_per_epoch = len(dm.ds_train)
scheduler = hydra.utils.instantiate(
scheduler_config.warmup_cosine, decay_steps=int(num_epochs * num_steps_per_epoch)
)
optimizer = optax.chain(optimizer, optax.scale_by_schedule(scheduler))
optimizer
GradientTransformationExtraArgs(init=<function chain.<locals>.init_fn at 0x15529cf904c0>, update=<function chain.<locals>.update_fn at 0x15529cf90160>)
def fft_mse_1D(y, yhat, flow=None, fhigh=None, reduction: str="mean"):
dims = y.shape
# FFT Transformation
y_f = jnp.fft.fftn(y)
yhat_f = jnp.fft.fftn(yhat)
print(y_f.shape[0])
if flow is None:
flow = 0
if fhigh is None:
fhigh = jnp.max(y_f.shape[0])-1
# # Subset the
# if len(dims) == 1:
# print("here")
# y_f = y_f[flow:fhigh]
# # y_f = y_f[slice(flow,fhigh,1)]
# y_f = jax.lax.dynamic_slice_in_dim(y_f, start_index=flow, slice_size=fhigh-flow)
# # yhat_f = yhat_f[slice(flow,fhigh,1)]
# # yhat_f = yhat_f[flow:fhigh]
# yhat_f = jax.lax.dynamic_slice_in_dim(yhat_f, start_index=flow, slice_size=fhigh-flow)
# elif len(dims) == 2:
# # y_f = y_f[flow:fhigh, flow:fhigh]
# # y_f = jax.lax.dynamic_slice_in_dim(y_f, start_index=[flow,flow], slice_size=[fhigh-flow,fhigh-flow])
# y_f = y_f[slice(flow,fhigh,1),slice(flow,fhigh,1)]
# # yhat_f = yhat_f[flow:fhigh, flow:fhigh]
# # yhat_f = jax.lax.dynamic_slice_in_dim(yhat_f, start_index=[flow,flow], slice_size=[fhigh-flow,fhigh-flow])
# yhat_f = yhat_f[slice(flow,fhigh,1),slice(flow,fhigh,1)]
# elif len(dims) == 3:
# y_f = y_f[flow:fhigh, flow:fhigh, flow:fhigh]
# yhat_f = yhat_f[flow:fhigh, flow:fhigh, flow:fhigh]
# elif len(dims) == 4:
# y_f = y_f[flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh]
# yhat_f = yhat_f[flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh]
# elif len(dims) == 5:
# y_f = y_f[flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh]
# yhat_f = yhat_f[flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh, flow:fhigh]
# else:
# raise ValueError(f"")
if reduction == "mean":
return jnp.abs(jnp.mean(y_f - yhat_f))
elif reduction == "sum":
return jnp.abs(jnp.sum(y_f - yhat_f))
else:
return jnp.abs(y_f - yhat_f)
Trainer Module#
import glob
import os
from pathlib import Path
from jejeqx._src.trainers.base import TrainerModule
from jejeqx._src.trainers.callbacks import wandb_model_artifact
from jejeqx._src.losses import psnr
class RegressorTrainer(TrainerModule):
def __init__(self, model, optimizer, **kwargs):
super().__init__(model=model, optimizer=optimizer, pl_logger=None, **kwargs)
def create_functions(self):
@eqx.filter_value_and_grad
def mse_loss(model, batch):
x, t, y = batch["spatial"], batch["temporal"], batch["data"]
pred = jax.vmap(model, in_axes=(0, 0))(x, t)
loss = jnp.mean((y - pred) ** 2)
return loss
def fft_mse_loss(model, batch):
x, t, y = batch["spatial"], batch["temporal"], batch["data"]
pred = jax.vmap(model, in_axes=(0,0))(x, t)
loss = fft_mse_1D(y=y, yhat=pred)
return loss
def train_step(state, batch):
loss, grads = mse_loss(state.params, batch)
state = state.update_state(state, grads)
psnr_loss = psnr(loss)
fft_loss = fft_mse_loss(state.params, batch)
metrics = {"loss": loss, "psnr": psnr_loss, "fft": fft_loss}
return state, loss, metrics
def eval_step(model, batch):
loss, _ = mse_loss(model, batch)
psnr_loss = psnr(loss)
return {"loss": loss, "psnr": psnr_loss}
def test_step(model, batch):
x, t = batch["spatial"], batch["temporal"]
out = jax.vmap(model, in_axes=(0, 0))(x, t)
loss, _ = mse_loss(model, batch)
psnr_loss = psnr(loss)
fft_loss = fft_mse_loss(model, batch)
metrics = {"loss": loss, "psnr": psnr_loss, "fft": fft_loss}
return out, metrics
def predict_step(model, batch):
x, t = batch["spatial"], batch["temporal"]
out = jax.vmap(model, in_axes=(0, 0))(x, t)
return out
return train_step, eval_step, test_step, predict_step
def on_training_end(
self,
):
if self.pl_logger:
save_dir = Path(self.log_dir).joinpath(self.save_name)
self.save_model(save_dir)
wandb_model_artifact(self)
self.pl_logger.finalize("success")
seed = 123
debug = False
enable_progress_bar = False
log_dir = "./"
trainer = RegressorTrainer(
model_mlp,
optimizer,
seed=seed,
debug=debug,
enable_progress_bar=enable_progress_bar,
log_dir=log_dir,
)
train_more = False
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
10000
401
CPU times: user 762 ms, sys: 7.84 ms, total: 770 ms
Wall time: 643 ms
{'fft': 0.48898881673812866,
'loss': 0.33336636424064636,
'psnr': 8.517776489257812}
xrda["ssh_pre"] = (("time", "lat", "lon"), dm.data_to_df(out).to_xarray().ssh.data)
try:
trainer.load_model("./checkpoints/checkpoint_model_mlp_ssh.ckpt")
except:
pass
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
CPU times: user 281 ms, sys: 11.2 ms, total: 292 ms
Wall time: 194 ms
{'fft': 0.007226995658129454,
'loss': 0.00013177159416954964,
'psnr': 82.71487426757812}
%%time
if train_more:
metrics = trainer.train_model(dm, num_epochs=num_epochs)
CPU times: user 9 µs, sys: 1e+03 ns, total: 10 µs
Wall time: 21.9 µs
if train_more:
trainer.save_model("./checkpoints/checkpoint_model_mlp_ssh.ckpt")
# trainer.save_model("./checkpoints/checkpoint_model_gabor_ssh.ckpt")
# trainer.save_state("checkpoint_state.ckpt")
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
{'fft': 0.007226995658129454,
'loss': 0.00013177159416954964,
'psnr': 82.71487426757812}
all_metrics = pd.DataFrame(
data=[["mlp", metrics["loss"], metrics["psnr"], metrics["fft"]]],
columns=["model", "MSE", "PSNR", "FFT"],
)
all_metrics
| model | MSE | PSNR | FFT | |
|---|---|---|---|---|
| 0 | mlp | 0.000132 | 82.714874 | 0.007227 |
dm.data_to_df(out).to_xarray().ssh
<xarray.DataArray 'ssh' (time: 1, lat: 201, lon: 201)>
array([[[ 0.5488428 , 0.54304546, 0.5354617 , ..., 0.5738971 ,
0.57589525, 0.5782987 ],
[ 0.54808766, 0.5422071 , 0.53459525, ..., 0.5805601 ,
0.5814417 , 0.5856366 ],
[ 0.55316144, 0.544704 , 0.5344246 , ..., 0.5869087 ,
0.58940536, 0.59366304],
...,
[-0.14343482, -0.14286944, -0.14328447, ..., -0.17531756,
-0.17495553, -0.17561796],
[-0.14795297, -0.14763284, -0.14773592, ..., -0.17243722,
-0.17006823, -0.16802837],
[-0.15249169, -0.15217172, -0.15206766, ..., -0.1639832 ,
-0.16152105, -0.15934393]]], dtype=float32)
Coordinates:
* time (time) float64 0.0
* lat (lat) float64 0.0 4.66e+03 9.318e+03 ... 8.708e+05 8.748e+05
* lon (lon) float64 0.0 5.56e+03 1.112e+04 ... 1.106e+06 1.112e+06xrda["ssh_mlp"] = (("time", "lat", "lon"), dm.data_to_df(out).to_xarray().ssh.data)
xrda
<xarray.Dataset>
Dimensions: (time: 1, lat: 201, lon: 201)
Coordinates:
* lon (lon) float64 -65.0 -64.95 -64.9 -64.85 ... -55.1 -55.05 -55.0
* lat (lat) float64 33.0 33.05 33.1 33.15 33.2 ... 42.85 42.9 42.95 43.0
* time (time) datetime64[ns] 2013-01-01
Data variables:
ssh (time, lat, lon) float64 0.5287 0.5287 0.523 ... -0.1701 -0.1701
ssh_pre (time, lat, lon) float32 -0.04211 -0.0421 ... -0.03984 -0.03981
ssh_mlp (time, lat, lon) float32 0.5488 0.543 0.5355 ... -0.1615 -0.1593ssh_fn_mlp = trainer.model
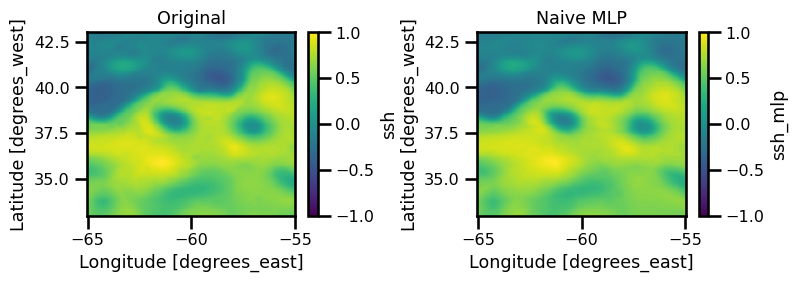
fig, ax = plt.subplots(ncols=2, figsize=(8, 3))
xrda.ssh.isel(time=0).plot.pcolormesh(ax=ax[0], cmap="viridis")
ax[0].set(title="Original")
xrda.ssh_mlp.isel(time=0).plot.pcolormesh(ax=ax[1], cmap="viridis")
ax[1].set(title="Naive MLP")
plt.tight_layout()
plt.show()
class FftMseLoss(object):
"""
loss function in Fourier space
June 2022, F.Alesiani
"""
def __init__(self, reduction='mean'):
super(FftMseLoss, self).__init__()
#Dimension and Lp-norm type are postive
self.reduction = reduction
def __call__(self, x, y, flow=None,fhigh=None, eps=1e-20):
num_examples = x.size()[0]
others_dims = x.shape[1:-2]
for d in others_dims:
assert (d>1), "we expect the dimension to be the same and greater the 1"
# print(others_dims)
dims = list(range(1,len(x.shape)-1))
xf = torch.fft.fftn(x,dim=dims)
yf = torch.fft.fftn(y,dim=dims)
if flow is None: flow = 0
if fhigh is None: fhigh = np.max(xf.shape[1:])
if len(others_dims) ==1:
xf = xf[:,flow:fhigh]
yf = yf[:,flow:fhigh]
if len(others_dims) ==2:
xf = xf[:,flow:fhigh,flow:fhigh]
yf = yf[:,flow:fhigh,flow:fhigh]
if len(others_dims) ==3:
xf = xf[:,flow:fhigh,flow:fhigh,flow:fhigh]
yf = yf[:,flow:fhigh,flow:fhigh,flow:fhigh]
if len(others_dims) ==4:
xf = xf[:,flow:fhigh,flow:fhigh,flow:fhigh,flow:fhigh]
yf = yf[:,flow:fhigh,flow:fhigh,flow:fhigh,flow:fhigh]
_diff = xf - yf
_diff = _diff.reshape(num_examples,-1).abs()**2
if self.reduction in ['mean']:
return torch.mean(_diff).abs()
if self.reduction in ['sum']:
return torch.sum(_diff).abs()
return _diff.abs()
if len(idxs) == 4: # 1D
nx = idxs[2]
pred_F = torch.fft.rfft(pred, dim=2)
target_F = torch.fft.rfft(target, dim=2)
_err_F = torch.sqrt(torch.mean(torch.abs(pred_F - target_F) ** 2, axis=0)) / nx * Lx
if len(idxs) == 5: # 2D
pred_F = torch.fft.fftn(pred, dim=[2, 3])
target_F = torch.fft.fftn(target, dim=[2, 3])
nx, ny = idxs[2:4]
_err_F = torch.abs(pred_F - target_F) ** 2
err_F = torch.zeros([nb, nc, min(nx // 2, ny // 2), nt]).to(device)
for i in range(nx // 2):
for j in range(ny // 2):
it = mt.floor(mt.sqrt(i ** 2 + j ** 2))
if it > min(nx // 2, ny // 2) - 1:
continue
err_F[:, :, it] += _err_F[:, :, i, j]
_err_F = torch.sqrt(torch.mean(err_F, axis=0)) / (nx * ny) * Lx * Ly
def fft_mse_1D_red():
return loss
xrda.ssh.data.shape
(1, 201, 201)
loss_mlp = fft_mse_1D(xrda.ssh.data.ravel(), xrda.ssh_mlp.data.ravel())
loss_pre = fft_mse_1D(xrda.ssh.data.ravel(), xrda.ssh_pre.data.ravel())
loss_mlp, loss_pre
(-4409.288-7171.893j) (15479.882+0j)
(-4409.288-7171.893j) (15479.882+0j)
(Array(8.443195, dtype=float32), Array(8403.085, dtype=float32))
ssh_f = np.fft.fftn(xrda.ssh.data)
ssh_mlp_f = np.fft.fftn(xrda.ssh_mlp.data)
ssh_pre_f = np.fft.fftn(xrda.ssh_pre.data)
ssh_f.shape, ssh_mlp_f.shape, ssh_pre_f.shape
((1, 201, 201), (1, 201, 201), (1, 201, 201))
flow = 0
fhigh = np.max(ssh_f.shape[1:])
flow, fhigh
(0, 201)
ssh_f_red = ssh_f[:, flow:fhigh, flow:fhigh]
ssh_mlp_f_red = ssh_mlp_f[:, flow:fhigh, flow:fhigh]
ssh_pre_f_red = ssh_pre_f[:, flow:fhigh, flow:fhigh]
ssh_f_red.shape, ssh_mlp_f_red.shape, ssh_pre_f_red.shape
((1, 201, 201), (1, 201, 201), (1, 201, 201))
diff_mlp = ssh_f_red - ssh_mlp_f_red
diff_pre = ssh_f_red - ssh_pre_f_red
np.abs(np.mean(diff_mlp))
0.020105064149144113
np.abs(np.mean(diff_pre))
0.5708498952434036
Random Fourier Features#
where \(\boldsymbol{\Omega}\) is a random matrix sampled from a Gaussian distribution.
So our final neural network with the additional basis function:
where \(\boldsymbol{\phi}(\cdot)\) is the learned basis network.
# load config
model_config = OmegaConf.load("./configs/model.yaml")
# instantiate
model_ffn = hydra.utils.instantiate(model_config.ffn)
# test output
out = model_ffn(x=x_init[0], t=t_init[0])
assert out.shape == y_init[0].shape
# test output (batched)
out_batch = jax.vmap(model_ffn, in_axes=(0, 0))(x_init, t_init)
assert out_batch.shape == y_init.shape
seed = 123
debug = False
enable_progress_bar = False
log_dir = "./"
trainer = RegressorTrainer(
model_ffn,
optimizer,
seed=seed,
debug=debug,
enable_progress_bar=enable_progress_bar,
log_dir=log_dir,
)
train_more = False
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
try:
trainer.load_model("./checkpoints/checkpoint_model_rff_ssh.ckpt")
except:
pass
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
%%time
if train_more:
metrics = trainer.train_model(dm, num_epochs=num_epochs)
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
if train_more:
trainer.save_model("./checkpoints/checkpoint_model_rff_ssh.ckpt")
all_metrics = pd.concat(
[
all_metrics,
pd.DataFrame(
data=[["rff", metrics["loss"], metrics["psnr"]]],
columns=["model", "MSE", "PSNR"],
),
]
)
all_metrics
xrda["ssh_rfe"] = (("time", "lat", "lon"), dm.data_to_df(out).to_xarray().ssh.data)
xrda["ssh_rfe"].attrs["standard_name"] = "Sea Surface Height"
ssh_fn_rff = trainer.model
fig, ax = plt.subplots(ncols=3, figsize=(12, 3))
xrda.ssh.isel(time=0).plot.pcolormesh(ax=ax[0], cmap="viridis")
ax[0].set(title="Original")
xrda.ssh_mlp.isel(time=0).plot.pcolormesh(ax=ax[1], cmap="viridis")
ax[1].set(title="Naive MLP")
xrda.ssh_rfe.isel(time=0).plot.pcolormesh(ax=ax[2], cmap="viridis")
ax[2].set(title="Fourier Features")
plt.tight_layout()
plt.show()
Custom Activation Functions#
SIREN
One of the most famous methods is the SIREN method. This replaces the standard activation function, \(\sigma\), with a sinusoidal function.
So our final neural network with the additional basis function:
where \(\boldsymbol{\phi}(\cdot)\) is the learned basis network.
# load config
model_config = OmegaConf.load("./configs/model.yaml")
# instantiate
model_siren = hydra.utils.instantiate(model_config.siren)
# test output
out = model_siren(x=x_init[0], t=t_init[0])
assert out.shape == y_init[0].shape
# test output (batched)
out_batch = jax.vmap(model_siren, in_axes=(0, 0))(x_init, t_init)
assert out_batch.shape == y_init.shape
seed = 123
debug = False
enable_progress_bar = False
log_dir = "./"
trainer = RegressorTrainer(
model_siren,
optimizer,
seed=seed,
debug=debug,
enable_progress_bar=enable_progress_bar,
log_dir=log_dir,
)
train_more = False
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
try:
trainer.load_model("./checkpoints/checkpoint_model_siren_ssh.ckpt")
except:
pass
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
%%time
if train_more:
metrics = trainer.train_model(dm, num_epochs=num_epochs)
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
all_metrics = pd.concat(
[
all_metrics,
pd.DataFrame(
data=[["siren", metrics["loss"], metrics["psnr"]]],
columns=["model", "MSE", "PSNR"],
),
]
)
all_metrics
if train_more:
trainer.save_model("./checkpoints/checkpoint_model_siren_ssh.ckpt")
xrda["ssh_siren"] = (("time", "lat", "lon"), dm.data_to_df(out).to_xarray().ssh.data)
xrda["ssh_siren"].attrs["standard_name"] = "Sea Surface Height"
ssh_fn_siren = trainer.model
fig, ax = plt.subplots(ncols=4, figsize=(16, 3))
xrda.ssh.isel(time=0).plot.pcolormesh(ax=ax[0], cmap="viridis")
ax[0].set(title="Original")
xrda.ssh_mlp.isel(time=0).plot.pcolormesh(ax=ax[1], cmap="viridis")
ax[1].set(title="Naive MLP")
xrda.ssh_rfe.isel(time=0).plot.pcolormesh(ax=ax[2], cmap="viridis")
ax[2].set(title="Fourier Features")
xrda.ssh_siren.isel(time=0).plot.pcolormesh(ax=ax[3], cmap="viridis")
ax[3].set(title="Siren")
plt.tight_layout()
plt.show()
Derived Variables#
# create function
ssh_fn: tp.Callable = ssh_fn_siren # ssh_fn_mlp # ssh_fn_rff #
Domain
Sea Surface Height
The proper sea surface height equation is given by:
Here, we learned a parameterized function, \(\boldsymbol{f_\theta}(\cdot)\), as a substitute for the true field. This is given by:
# create FAST batched function
ssh_fn_batch: tp.Callable = jax.vmap(ssh_fn, in_axes=(0, 0))
# make it FAST!
ssh_fn_batch: tp.Callable = jax.jit(ssh_fn_batch, backend="cpu")
# predict on the batch of coordinates
ssh: Array = ssh_fn_batch(dm.ds_test[:]["spatial"], dm.ds_test[:]["temporal"])
def make_vmap_st(f):
return jax.vmap(f, in_axes=(0, 0))
def make_jit_cpu(f):
return jax.jit(f, backend="cpu")
# create data array
xr_ssh: xr.Dataset = dm.data_to_df(ssh).to_xarray().ssh
# quick plot
xr_ssh.isel(time=0).plot.pcolormesh(cmap="viridis")
INTERLUDE: We can query wherever we want!
num_x = 500
num_y = 500
num_t = 1
x_coords = np.linspace(-0.5, 0.5, num_x)
y_coords = np.linspace(-0.5, 0.5, num_y)
t_coords = np.linspace(0, 0, num_t)
import einops
# create coordinates
XYT = np.meshgrid(x_coords, y_coords, t_coords, indexing="ij")
XYT = np.stack(XYT, axis=-1)
# XY, T = XYT[..., :-1], XYT[..., -1]
XYT = einops.rearrange(XYT, "H W T D -> (H W T) D")
XY, T = XYT[..., :-1], XYT[..., -1]
XY.shape, T.shape
out = ssh_fn_batch(XY, T)
print(out.shape)
out = einops.rearrange(out, "(H W T) D -> H W T D", H=num_x, W=num_y, T=num_t)
out.shape
%matplotlib inline
plt.scatter(XY[..., 1], XY[..., 0], c=out.squeeze())
Stream Function#
from jejeqx._src.transforms.xarray.geostrophic import calculate_coriolis
from metpy.constants import earth_gravity
f0 = calculate_coriolis(xrda.lat)
print(f"Coriolis Parameter:", f0.data)
print(f"Earth Gravity:", earth_gravity)
f0: Array = jnp.asarray(calculate_coriolis(xrda.lat).data.magnitude)
g: Array = jnp.asarray(earth_gravity.magnitude)
def create_streamfn(f: tp.Callable, f0: float = 1e-5, g: float = 9.81) -> tp.Callable:
def sfn(x: Array, t: Array) -> Array:
return (g / f0) * f(x, t)
return sfn
psi_fn = create_streamfn(ssh_fn)
# create FAST Batched Function
psi_fn_batch = make_jit_cpu(make_vmap_st(psi_fn))
# make predictions
psi = psi_fn_batch(dm.ds_test[:]["spatial"], dm.ds_test[:]["temporal"])
# create xarray dataset
xrda["psi"] = (g / f0) * xrda.ssh
xrda["psi_ad"] = (("time", "lat", "lon"), dm.data_to_df(psi).to_xarray().ssh.data)
# demo plot
xrda["psi_ad"].isel(time=0).plot.pcolormesh(cmap="viridis")
Velocities#
def create_gradient_fn(f: tp.Callable) -> tp.Callable:
def fn(x: Array, t: Array) -> Array:
return jax.jacfwd(f)(x, t).squeeze()
return fn
def uv_velocity(grad_psi: Array) -> tp.Tuple[Array, Array]:
dpsi_x, dpsi_y = jnp.split(grad_psi, 2, axis=-1)
u = -dpsi_y
v = dpsi_x
return u, v
grad_psi_fn = create_gradient_fn(psi_fn)
grad_psi_fn_batched = make_jit_cpu(make_vmap_st(grad_psi_fn))
# make predictions
grad_psi = grad_psi_fn_batched(dm.ds_test[:]["spatial"], dm.ds_test[:]["temporal"])
# parse to get velocity
u, v = uv_velocity(grad_psi)
# create xarray dataset
xrda["u_ad"] = (("time", "lat", "lon"), dm.data_to_df(u).to_xarray().ssh.data)
xrda["v_ad"] = (("time", "lat", "lon"), dm.data_to_df(v).to_xarray().ssh.data)
xrda["ke_ad"] = np.hypot(xrda["u_ad"], xrda["v_ad"])
fig, ax = plt.subplots()
xrda["ke_ad"].isel(time=0).plot.pcolormesh(ax=ax, cmap="YlGnBu_r")
plt.tight_layout()
plt.show()
Finite Difference#
So we can also do this in discrete space as well. Meaning, we can take the derivative of the field we are interested in. $\( \frac{\partial \psi}{\partial x}= D[\psi](\vec{\mathbf{x}}) \)$
We have many different types, e.g. central difference, forwards difference and backwards difference.
import jejeqx._src.transforms.xarray.geostrophic as geocalc
xrda = geocalc.calculate_velocities_sf(xrda, "psi")
xrda = geocalc.calculate_kinetic_energy(xrda, ["u", "v"])
xrda
fig, ax = plt.subplots(ncols=2, figsize=(10, 4))
xrda["ke_ad"].isel(time=0).plot.pcolormesh(ax=ax[0], cmap="YlGnBu_r")
ax[0].set(title="Auto-Differentiation")
xrda["ke"].isel(time=0).plot.pcolormesh(ax=ax[1], cmap="YlGnBu_r")
ax[1].set(title="Finite Difference")
plt.tight_layout()
plt.show()
Relative Vorticity#
Somtimes called the vertical vorticity.
Note that the u,v velocities can be calculated from the stream function as
So plugging these into the equation, we get:
We can also calculate a normalized version
Note: This is closely related to the geostrophic eqns:
def create_laplacian_fn(f: tp.Callable) -> tp.Callable:
def fn(x: Array) -> Array:
# return jax.jacfwd(jax.jacrev(f))(x)
H = jax.hessian(f)
L = jnp.diagonal(H(x)[0])
return jnp.sum(L, keepdims=True)
return fn
rvort_fn = create_laplacian_fn(psi_fn)
rvort_fn(x_init[0])
rvort_fn_batched = jax.jit(jax.vmap(rvort_fn, in_axes=(0, 0)), backend="cpu")
rvort = rvort_fn_batched(dm.ds_test[:]["spatial"], dm.ds_test[:]["temporal"])
# create xarray dataset
xrda["rvort"] = dm.data_to_df(rvort).to_xarray().ssh
import finitediffx as fdx
method = "central"
order = 1
step_size = 1
accuracy = 1
dv_dx = dfdx(xrda["v_fd"].data)
du_dy = dfdy(xrda["u_fd"].data)
xrda["rvort_fd"] = (("time", "lat", "lon"), dv_dx - du_dy)
fig, ax = plt.subplots(ncols=2, figsize=(10, 4))
xrda["rvort"].isel(time=0).plot.pcolormesh(ax=ax[0], cmap="RdBu_r")
ax[0].set(title="Auto-Differentiation")
xrda["rvort_fd"].isel(time=0).plot.pcolormesh(ax=ax[1], cmap="RdBu_r")
ax[1].set(title="Finite Difference")
plt.tight_layout()
plt.show()
import common_utils as cutils
ds_rfe = cutils.calculate_physical_quantities(xrda.ssh_rfe)
ds_natl60 = cutils.calculate_physical_quantities(xrda.ssh)
ds_mlp = cutils.calculate_physical_quantities(xrda.ssh_mlp)
ds_siren = cutils.calculate_physical_quantities(xrda.ssh_siren)
def rmse_da(da, da_ref, dim):
return ((da - da_ref) ** 2).mean(dim=dim) ** 0.5
def nrmse_da(da, da_ref, dim):
rmse = rmse_da(da=da, da_ref=da_ref, dim=dim)
std = (da_ref**2).mean(dim=dim) ** 0.5
return 1.0 - (rmse / std).data.magnitude
import pandas as pd
dims = ["lat", "lon"]
results_df = pd.DataFrame()
for imodel, iname in zip([ds_mlp, ds_rfe, ds_siren], ["MLP", "RFE", "SIREN"]):
for ivar in imodel:
error = nrmse_da(imodel[ivar], ds_natl60[ivar], dims)
ires_df = pd.DataFrame(
data=[[iname, ivar, error.item()]],
columns=[
"model",
"variable",
"nrmse",
],
)
results_df = pd.concat([ires_df, results_df.loc[:]], axis=0)
results_df.head()
results_df.loc[results_df["variable"] == "ke"].sort_values("nrmse")
fig, ax = cutils.plot_analysis_vars(
[
ds_natl60.isel(time=0),
ds_mlp.isel(time=0),
ds_rfe.isel(time=0),
ds_siren.isel(time=0),
],
figsize=(30, 30),
)
# fig.suptitle("NATL60 | MLP | RFE | SIREN"),
plt.show()
#
ds_psd_natl60 = cutils.calculate_isotropic_psd(ds_natl60)
ds_psd_rfe = cutils.calculate_isotropic_psd(ds_rfe)
ds_psd_mlp = cutils.calculate_isotropic_psd(ds_mlp)
ds_psd_siren = cutils.calculate_isotropic_psd(ds_siren)
fig, ax = cutils.plot_analysis_psd_iso(
[ds_psd_natl60, ds_psd_mlp, ds_psd_rfe, ds_psd_siren],
["NATL60", "MLP", "RFE", "SIREN"],
)
plt.show()
ds_rfe_scores = cutils.calculate_isotropic_psd_score(ds_rfe, ds_natl60)
ds_mlp_scores = cutils.calculate_isotropic_psd_score(ds_mlp, ds_natl60)
ds_siren_scores = cutils.calculate_isotropic_psd_score(ds_siren, ds_natl60)
import pandas as pd
results_df = pd.DataFrame(
columns=["model", "variable", "wavelength [km]", "wavelength [degree]"]
)
for iscore, imodel in zip(
[ds_mlp_scores, ds_rfe_scores, ds_siren_scores], ["MLP", "RFE", "SIREN"]
):
for ivar in iscore:
resolved_spatial_scale = iscore[ivar].attrs["resolved_scale_space"] / 1e3
ires_df = pd.DataFrame(
data=[[imodel, ivar, resolved_spatial_scale, resolved_spatial_scale / 111]],
columns=["model", "variable", "wavelength [km]", "wavelength [degree]"],
)
results_df = pd.concat([ires_df, results_df.loc[:]], axis=0)
results_df.head()
results_df.loc[results_df["variable"] == "strain"].sort_values("wavelength [km]")
cutils.plot_analysis_psd_iso_score(
[ds_mlp_scores, ds_rfe_scores, ds_siren_scores],
["MLP", "RFE", "SIREN"],
["b", "orange", "green"],
)
plt.show()

