OSSE SWOT#
import autoroot
import jax
import jax.numpy as jnp
import jax.scipy as jsp
import jax.random as jrandom
import numpy as np
import numba as nb
import equinox as eqx
import kernex as kex
import finitediffx as fdx
import diffrax as dfx
import xarray as xr
import metpy
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm.notebook import tqdm, trange
from jaxtyping import Float, Array, PyTree, ArrayLike
import wandb
from omegaconf import OmegaConf
import hydra
from sklearn import pipeline
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from jejeqx._src.transforms.dataframe.spatial import Spherical2Cartesian
from jejeqx._src.transforms.dataframe.temporal import TimeDelta
from jejeqx._src.transforms.dataframe.scaling import MinMaxDF
sns.reset_defaults()
sns.set_context(context="talk", font_scale=0.7)
# Ensure TF does not see GPU and grab all GPU memory.
%env XLA_PYTHON_CLIENT_PREALLOCATE=false
jax.config.update("jax_enable_x64", False)
%matplotlib inline
%load_ext autoreload
%autoreload 2
Recap Formulation#
We are interested in learning non-linear functions \(\boldsymbol{f}\).
where the \(\boldsymbol{\phi}(\cdot)\) is a basis function. Neural Fields typically try to learn this basis funciton via a series of composite functions of the form
Problems#
Here, we will demonstrate a problem that a naive network has.
Sparse Observations#
In the previous examples, we were demonstrating how NerFs perform when we have some clean simulation. However, in many real problems, we do not have access to such clean
For this example, we are going to look at the case when we have very sparse observations: as in the case with satellite altimetry data like SWOT. In this case
from dataclasses import dataclass, field
from typing import List, Dict, Optional
files = [
"/gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/work_eman/ml_ready/swot1nadir5.nc",
# "/gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/work_eman/ml_ready/nadir4.nc",
]
@dataclass
class SSHDM:
_target_: str = "jejeqx._src.datamodules.coords.AlongTrackDM"
paths: List[str] = field(default_factory=lambda: files)
batch_size: int = 10_000
shuffle: bool = True
train_size: float = 0.80
decode_times: bool = True
spatial_coords: List = field(default_factory=lambda: ["lat", "lon"])
temporal_coords: List = field(default_factory=lambda: ["time"])
variables: List = field(default_factory=lambda: ["ssh_obs"])
# spatial transform
spatial_transforms = Pipeline(
[
("cartesian3d", Spherical2Cartesian(radius=1.0, units="degrees")),
("spatialminmax", MinMaxDF(["x", "y", "z"], -1, 1)),
]
)
temporal_transforms = Pipeline(
[
("timedelta", TimeDelta("2012-10-01", 1, "s")),
("timeminmax", MinMaxDF(["time"], -1, 1)),
]
)
select = {"time": slice("2012-10-01", "2012-12-02")}
config_dm = OmegaConf.structured(SSHDM())
dm = hydra.utils.instantiate(
config_dm,
select=select,
spatial_transform=spatial_transforms,
temporal_transform=temporal_transforms,
)
dm.setup()
init = dm.ds_train[:32]
x_init, t_init, y_init = init["spatial"], init["temporal"], init["data"]
x_init.min(), x_init.max(), x_init.shape, t_init.min(), t_init.max(), t_init.shape
len(dm.ds_train)
xrda_obs = dm.load_xrds()
xrda_obs
# fig, ax = plt.subplots(ncols=1, figsize=(5,4))
# xrda_obs.ssh_obs.isel(time=1).plot.pcolormesh(ax=ax, cmap="viridis")
# ax.set(title="Original")
# plt.tight_layout()
# plt.show()
# import geoviews as gv
# import geoviews.feature as gf
# from cartopy import crs
# gv.extension('bokeh', 'matplotlib')
# xrda_obs
# dataset = gv.Dataset(xrda_obs)
# ensemble1 = dataset.to(gv.Image, ['lon', 'lat'], "ssh_obs")
# gv.output(ensemble1.opts(cmap='viridis', colorbar=True, fig_size=200, backend='matplotlib') * gf.coastline(),
# backend='matplotlib')
# lr = 5e-3
# num_epochs = 5_000
# num_steps_per_epoch = len(dm.ds_train)
# @dataclass
# class FoxDataModule:
# _target_: str = "jejeqx._src.datamodules.image.ImageFox"
# batch_size: int = 10_000
# train_size: float = 0.5
# shuffle: bool = False
# split_method: str = "even"
# resize: int = 4
# @dataclass
# class Training:
# num_epochs: int = 2_000
# @dataclass
# class Model:
# _target_: str = "jejeqx._src.nets.nerfs.siren.SirenNet"
# in_size: int = 2
# out_size: int = 3
# width_size: int = 128
# depth: int = 5
# @dataclass
# class Optimizer:
# _target_: str = "optax.adam"
# learning_rate: float = lr
# @dataclass
# class Scheduler:
# _target_: str = "optax.warmup_cosine_decay_schedule"
# init_value: float = 0.0
# peak_value: float = lr
# warmup_steps: int = 100
# decay_steps: int = int(num_epochs * num_steps_per_epoch)
# end_value: float = 0.01 * lr
# @dataclass
# class Config:
# datamodule: FoxDataModule = FoxDataModule()
# model: Model = Model()
# optimizer: Optimizer = Optimizer()
# scheduler: Scheduler = Scheduler()
# num_epochs: int = 2_000
# import optax
# config = Config()
# config = OmegaConf.structured(Config())
# # initialize datamodule
# dm = hydra.utils.instantiate(config.datamodule)
# dm.setup()
# # initialize optimizer
# optimizer = hydra.utils.instantiate(config.optimizer)
# # initialize scheduler
# num_steps_per_epoch = len(dm.ds_train)
# decay_steps = int(num_steps_per_epoch * config.num_epochs)
# schedule_fn = hydra.utils.instantiate(config.scheduler, decay_steps=decay_steps)
# # initialize optimizer + scheduler
# optimizer = optax.chain(optimizer, optax.scale_by_schedule(schedule_fn))
# ibatch = next(iter(dm.train_dataloader()))
# print(ibatch[0].shape, ibatch[1].shape, type(ibatch[0]))
Model#
The input data is a coordinate vector, \(\mathbf{x}_\phi\), of the image coordinates.
where \(D_\phi = [\text{x}, \text{y}]\). So we are interested in learning a function, \(\boldsymbol{f}\), such that we can input a coordinate vector and output a scaler/vector value of the pixel value.
SIREN Layer#
where \(\mathbf{s}\) is the modulation
Siren Model#
!ls /gpfswork/rech/cli/uvo53rl/checkpoints/nerfs/siren/swot1nadir5/
import joblib
model_saved = "pretrained" # "scratch" #
model_config_file = f"/gpfswork/rech/cli/uvo53rl/checkpoints/nerfs/siren/swot1nadir5/{model_saved}/config.pkl"
checkpoint_file = f"/gpfswork/rech/cli/uvo53rl/checkpoints/nerfs/siren/swot1nadir5/{model_saved}/checkpoint_model.ckpt"
old_config = joblib.load(model_config_file)
model = hydra.utils.instantiate(old_config["model"])
from jejeqx._src.nets.nerfs.ffn import RFFLayer
model_name = "rff"
model = eqx.nn.Sequential(
[
RFFLayer(in_dim=4, num_features=256, out_dim=256, key=jrandom.PRNGKey(42)),
RFFLayer(in_dim=256, num_features=256, out_dim=256, key=jrandom.PRNGKey(123)),
RFFLayer(in_dim=256, num_features=256, out_dim=256, key=jrandom.PRNGKey(23)),
RFFLayer(in_dim=256, num_features=256, out_dim=256, key=jrandom.PRNGKey(81)),
RFFLayer(in_dim=256, num_features=256, out_dim=1, key=jrandom.PRNGKey(32)),
]
)
# check output of models
out = jax.vmap(model)(jnp.hstack([x_init, t_init]))
assert out.shape == y_init.shape
model_name = "siren"
@dataclass
class Key:
_target_: str = "jax.random.PRNGKey"
seed: int = 123
@dataclass
class SirenBasis:
_target_: str = "jejeqx._src.nets.nerfs.siren.SirenNet"
in_size: int = 4
out_size: int = 256
width_size: int = 256
depth: int = 5
key: Key = Key()
@dataclass
class LinearModel:
_target_: str = "equinox.nn.Linear"
in_features: int = 256
out_features: int = 1
use_bias: bool = True
key: Key = Key()
@dataclass
class NerFModel:
_target_: str = "jejeqx._src.nets.nerfs.base.NerF"
# basis_net: RFFBasis = RFFBasis()
basis_net: SirenBasis = SirenBasis()
network: LinearModel = LinearModel()
# initialize model
model_config = OmegaConf.structured(NerFModel())
model = hydra.utils.instantiate(model_config)
# check output of models
out = jax.vmap(model)(jnp.hstack([x_init, t_init]))
assert out.shape == y_init.shape
Optimizer (+ Learning Rate)#
For this, we will use a simple adam optimizer with a learning_rate of 1e-4. From many studies, it appears that a lower learning rate works well with this methods because there is a lot of data. In addition, a bigger batch_size is also desireable. We will set the num_epochs to 1_000 which should be good enough for a single image. Obviously more epochs and a better learning rate scheduler would result in better results but this will be sufficient for this demo.
Scheduler#
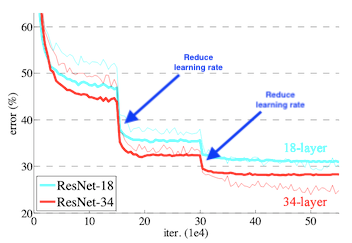
We will use a simple learning rate scheduler - reduce_lr_on_plateau. This will automatically reduce the learning rate as the validation loss stagnates. It will ensure that we really squeeze out as much performance as possible from our models during the training procedure.We start with a (relatively) high learning_rate of 1e-4 so we will set the patience to 5 epochs. So if there is no change in with every epoch, we decrease the learning rate by a factor of 0.1.
This is a rather crude (but effective) method but it tends to work well in some situations. A better method might be the cosine_annealing method or the exponential_decay method. See other examples.
import optax
num_epochs = 1_000
@dataclass
class Optimizer:
_target_: str = "optax.adam"
learning_rate: float = 1e-4
# @dataclass
# class Scheduler:
# _target_: str = "optax.warmup_exponential_decay_schedule"
# init_value: float = 0.0
# peak_value: float = 1e-2
# warmup_steps: int = 100
# end_value: float = 1e-5
# decay_rate: float = 0.1
# FINETUNE!
@dataclass
class Scheduler:
_target_: str = "optax.warmup_cosine_decay_schedule"
init_value: float = 0.0
peak_value: float = 1e-2
warmup_steps: int = 500
end_value: float = 1e-6
scheduler_config = OmegaConf.structured(Scheduler())
optim_config = OmegaConf.structured(Optimizer())
optimizer = hydra.utils.instantiate(optim_config)
# num_steps_per_epoch = len(dm.ds_train)
# scheduler = hydra.utils.instantiate(
# scheduler_config,
# decay_steps=int(num_epochs * num_steps_per_epoch)
# )
# optimizer = optax.chain(optimizer, optax.scale_by_schedule(scheduler))
Trainer Module#
import glob
import os
from pathlib import Path
from jejeqx._src.trainers.base import TrainerModule
from jejeqx._src.trainers.callbacks import wandb_model_artifact
from jejeqx._src.losses import psnr
class RegressorTrainer(TrainerModule):
def __init__(self, model, optimizer, **kwargs):
super().__init__(model=model, optimizer=optimizer, pl_logger=None, **kwargs)
@property
def model(self):
return self.state.params
@property
def model_batch(self):
return jax.vmap(self.state.params, in_axes=(0, 0))
def create_functions(self):
@eqx.filter_value_and_grad
def mse_loss(model, batch):
x, t, y = batch["spatial"], batch["temporal"], batch["data"]
# pred = jax.vmap(model, in_axes=(0,0))(x, t)
pred = jax.vmap(model)(jnp.hstack([x, t]))
loss = jnp.mean((y - pred) ** 2)
return loss
def train_step(state, batch):
loss, grads = mse_loss(state.params, batch)
state = state.update_state(state, grads)
psnr_loss = psnr(loss)
metrics = {"loss": loss, "psnr": psnr_loss}
return state, loss, metrics
def eval_step(model, batch):
loss, _ = mse_loss(model, batch)
psnr_loss = psnr(loss)
return {"loss": loss, "psnr": psnr_loss}
def test_step(model, batch):
x, t, y = batch["spatial"], batch["temporal"], batch["data"]
pred = jax.vmap(model)(jnp.hstack([x, t]))
loss, _ = mse_loss(model, batch)
psnr_loss = psnr(loss)
return pred, {"loss": loss, "psnr": psnr_loss}
def predict_step(model, batch):
x, t = batch["spatial"], batch["temporal"]
pred = jax.vmap(model)(jnp.hstack([x, t]))
return pred
return train_step, eval_step, test_step, predict_step
def on_training_end(
self,
):
if self.pl_logger:
save_dir = Path(self.log_dir).joinpath(self.save_name)
self.save_model(save_dir)
wandb_model_artifact(self)
self.pl_logger.finalize("success")
seed = 123
debug = False
enable_progress_bar = False
log_dir = "./"
trainer = RegressorTrainer(
model,
optimizer,
seed=seed,
debug=debug,
enable_progress_bar=enable_progress_bar,
log_dir=log_dir,
)
train_more = True
save_more = True
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
trainer.load_model(checkpoint_file)
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
%%time
# metrics = trainer.train_model(dm, num_epochs=num_epochs)
%%time
out, metrics = trainer.test_model(dm.test_dataloader())
metrics
Evaluation#
We will predict the whole dataset at the full resolution available for the same time period.
01-June-2013 :--> 15-June-2013
from dataclasses import dataclass, field
from typing import List, Dict
@dataclass
class SSHDMEVAL:
_target_: str = "jejeqx._src.datamodules.coords.EvalCoordDM"
paths: str = "/gpfswork/rech/yrf/commun/data_challenges/dc20a_osse/test/dc_ref/NATL60-CJM165_GULFSTREAM*"
batch_size: int = 10_000
shuffle: bool = False
train_size: float = 0.80
decode_times: bool = True
evaluation: bool = True
spatial_coords: List = field(default_factory=lambda: ["lat", "lon"])
temporal_coords: List = field(default_factory=lambda: ["time"])
variables: List = field(default_factory=lambda: ["sossheig"])
coarsen: Dict = field(default_factory=lambda: {"lon": 2, "lat": 2})
resample: str = "1D"
%%time
select = {"time": slice("2012-10-22", "2012-12-02")}
config_dm = OmegaConf.structured(SSHDMEVAL())
dm_eval = hydra.utils.instantiate(
config_dm,
select=select,
spatial_transform=dm.spatial_transform,
temporal_transform=dm.temporal_transform,
)
dm_eval.setup()
print(f"Num Points: {len(dm_eval.ds_test):,}")
%%time
xrda = dm_eval.load_xrds()
%%time
out, metrics = trainer.test_model(dm_eval.test_dataloader())
metrics
xrda["ssh_rff"] = dm_eval.data_to_df(out).to_xarray().sossheig
import common_utils as cutils
ds_rff = cutils.calculate_physical_quantities(xrda.ssh_rff)
ds_natl60 = cutils.calculate_physical_quantities(xrda.sossheig)
fig, ax = cutils.plot_analysis_vars(
[
ds_natl60.isel(time=15),
ds_rff.isel(time=15),
]
)
plt.show()
ds_psd_natl60 = cutils.calculate_isotropic_psd(ds_natl60)
ds_psd_rff = cutils.calculate_isotropic_psd(ds_rff)
fig, ax = cutils.plot_analysis_psd_iso(
[
ds_psd_natl60,
ds_psd_rff,
],
[
"NATL60",
"RFE",
],
)
plt.show()
ds_psd_scores = cutils.calculate_isotropic_psd_score(ds_rff, ds_natl60)
cutils.plot_analysis_psd_iso_score([ds_psd_scores], ["SIREN"], ["k"])
plt.show()
for ivar in ds_psd_scores:
resolved_spatial_scale = ds_psd_scores[ivar].attrs["resolved_scale_space"] / 1e3
print(f"Wavelength [km]: {resolved_spatial_scale:.2f} [{ivar.upper()}]")
print(f"Wavelength [degree]: {resolved_spatial_scale/111:.2f} [{ivar.upper()}]")
ds_psd_natl60 = cutils.calculate_spacetime_psd(ds_natl60)
ds_psd_rff = cutils.calculate_spacetime_psd(ds_rff)
fig, ax = cutils.plot_analysis_psd_spacetime(
[
ds_psd_natl60,
ds_psd_rff,
],
[
"NATL60",
"RFE",
],
)
plt.show()
ds_psd_rff = cutils.calculate_spacetime_psd_score(ds_rff, ds_natl60)
for ivar in ds_psd_rff:
resolved_spatial_scale = ds_psd_rff[ivar].attrs["resolved_scale_space"] / 1e3
print(f"Resolved Scale [km]: {resolved_spatial_scale:.2f} [{ivar.upper()}]")
resolved_temporal_scale = ds_psd_rff[ivar].attrs["resolved_scale_time"]
print(f"Resolved Scale [days]: {resolved_temporal_scale:.2f} [{ivar.upper()}]")
_ = cutils.plot_analysis_psd_spacetime_score([ds_psd_rff], ["rff"])

